Difference between revisions of "Main Page"
(→Experimental data) |
(Publications) |
||
| (222 intermediate revisions by 10 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
| − | + | '''Quick links''' | |
| − | + | Direct link to the page describing the [[NMReDATA tag format|format of the NMREDATA tags (version 1.0/1.1)]] and [[NMReDATA tag format 2.0|format of the NMREDATA tags (version 2.0)]]. | |
| − | |||
| − | [[ | + | List of [[compatible software|compatible software and webtools]] |
| − | + | Tentative instruction for [[Submission NMReDATA|journal submission of NMReDATA]]. | |
| − | + | Link to the [https://github.com/NMReDATAInitiative GitHub page] of the NMReDATA Initiative. | |
| − | = | + | = Introduction = |
| − | + | The [http://www.nmredata.org/partners.html NMReDATA working group] decided to include data extracted from NMR spectra of small molecules in [[sdf files| SDF files]] using SD tags. | |
| − | |||
| − | + | Link to the [https://onlinelibrary.wiley.com/doi/abs/10.1002/mrc.4737 MRC article] on the NMReDATA format. | |
| − | + | An important task of the group is to define the format of the content of the "<NMREDATA_...>" tags. [[NMReDATA tag format|More details here!]]. | |
| − | + | The version 1.0 has been decided in September at the "Round table" of the Smash 2017 conference at Baveno, Italy. | |
| − | + | The SDF file alone (that is without the spectra) cannot be used to verify that the assignment corresponds to the spectra. It is therefore important to always have the spectra with the SDF file! We call ''"NMR Record"'' the combination of the spectra and the SDF file. | |
| − | |||
| − | + | A 1.1 version was released to fix a problem in 1.0. | |
| + | Version 2.0 adds extensions and was released after the [[Symposium2019|'''1st NMReDATA symposium''']] (Sept. 16, Porto, Portugal). | ||
| − | = | + | = NMR records = |
| + | <!-- [[commented_example_nmredata|Examples of commented and simplified NMREDATA tags ]] --> | ||
| − | + | We call "NMR record", a folder (or .zip file including the folder) or a database record including: | |
| − | |||
| − | + | 1) All the NMR spectra (including FID, acquisition and processing parameters). The format of these data is as produced by the manufacturer of the instrument which acquired the data. That means that software generating the data either has these crude data available or it will ask the user to point to the crude data in order to include them in the ''NMR record''. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | 2) The SDF file(s) including the NMReDATA (''.nmredata.sdf'' file) | |
| − | |||
| − | |||
| − | + | [[File:nmr_record.png|600px|center|NMR record]] | |
| + | A more detailed picture (see [[File:test-2.pdf|600px|center|pictorial representation of NMR record and example of SDF file]]) was presented in the [http://nmredata.org/euromar_2017_v5_optimized.pdf poster] presented in July at the Euromar 2017. '''Note:''' The NMREDATA tag "SIGNALS" was renamed "ASSIGNMENT" in Version 0.98. | ||
| − | |||
| − | + | NMR records will be [http://onlinelibrary.wiley.com/doi/10.1002/mrc.4631/full requested by ''Magnetic Resonance in Chemistry''] from 2018 on. The editors of software (ADC/Labs, Bruker, cheminfo, Mestrelab) are able to grenerate MNR records. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Records will be either analysed on web pages, or downloaded, and the nmredata.sdf file opened by the software which will access automatically to the associated spectra. | |
| − | The | + | The full description can be found [[NMReDATA tag format|in the NMReDATA tag format page]]. |
| − | + | An example ('''probably obsolete''') of .nmredata.sdf files with the spectra can be found [https://www.dropbox.com/sh/hu0qudy2bt56ix0/AACc8UiUoeEskSDVhYnP-cZna?dl=0 here] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | A better source is the [https://github.com/NMReDATAInitiative/Examples-of-NMR-records GitHub example page] of the [https://github.com/nmredatainitiative GitHub page of the Initiative]. | |
| − | + | == Mixtures of compounds == | |
| − | + | When more than one compound is present in a sample, multiple .sdf are produced and called compound1.nmredata.sdf, compound2.nmredata.sdf, etc. | |
| − | + | = Current version of the format of NMReDATA = | |
| − | + | The format can be found here : [[NMReDATA tag format]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Various examples of NMReDATA files can be found in the [https://github.com/NMReDATAInitiative/Examples-of-NMR-records github repository]. | |
| − | + | = Database = | |
| − | + | The NMReDATA Inititiative define the format and rules for hosting and provide data, but hosting data is not part of our mission. | |
| − | + | Discussion on [[database_policy | about database providers ]] | |
| − | |||
| − | |||
| − | |||
| + | = Versions = | ||
| + | ==Changes to Versions 1.1 == | ||
| − | + | *[[end-of-line | Addition of backslash]] at the end of the line of NMReDATA tags. | |
| − | = | + | ==Changes to Versions 2.0 == |
| − | + | *Possibility to include Jcamp spectra on top of original data from the instrument. | |
| + | *Possibility to add a 3D structure (on top of the "flat" 2D structure). | ||
| + | *Possibility to add author and institution information (to facilitate integration of these metadata in repositories). | ||
| − | + | For details, see [[NMReDATA_tag_format_2.0|the specification]]. | |
| − | 2) | + | ==Changes to Versions 2.1 (future version)== |
| + | <span style="color:#FF8C00"> ''Future version of the format! Suggestions are welcome! '' </span> | ||
| + | *Possibility to include [[Jassign|J-coupling assignment data]] (coupling network and J-graph data). | ||
| − | + | Discussion and complete list on [[Future version| future versions of the NMReDATA format ]] | |
| − | + | = Jmol integration in this wiki = | |
| + | NMReDATA is being integrated with [[Jmol | Jmol/JspecView ]] | ||
| − | + | = Publications = | |
| + | * NMReDATA, a standard to report the NMR assignment and parameters of organic compounds. M. Pupier et al. (2018) Magnetic Resonance in Chemistry 56:703-715. doi: [https://doi.org/10.1002/mrc.4737 10.1002/mrc.4737] PMID: 29656574 | ||
| + | * NMReDATA: Tools and applications. S. Kuhn et al. (2021) Magnetic Resonance in Chemistry 59:792-803. doi: [https://doi.org/10.1002/mrc.5146 10.1002/mrc.5146] PMID: 33729627 | ||
Latest revision as of 21:42, 17 February 2024
Quick links
Direct link to the page describing the format of the NMREDATA tags (version 1.0/1.1) and format of the NMREDATA tags (version 2.0).
List of compatible software and webtools
Tentative instruction for journal submission of NMReDATA.
Link to the GitHub page of the NMReDATA Initiative.
Contents
Introduction
The NMReDATA working group decided to include data extracted from NMR spectra of small molecules in SDF files using SD tags.
Link to the MRC article on the NMReDATA format.
An important task of the group is to define the format of the content of the "<NMREDATA_...>" tags. More details here!.
The version 1.0 has been decided in September at the "Round table" of the Smash 2017 conference at Baveno, Italy.
The SDF file alone (that is without the spectra) cannot be used to verify that the assignment corresponds to the spectra. It is therefore important to always have the spectra with the SDF file! We call "NMR Record" the combination of the spectra and the SDF file.
A 1.1 version was released to fix a problem in 1.0.
Version 2.0 adds extensions and was released after the 1st NMReDATA symposium (Sept. 16, Porto, Portugal).
NMR records
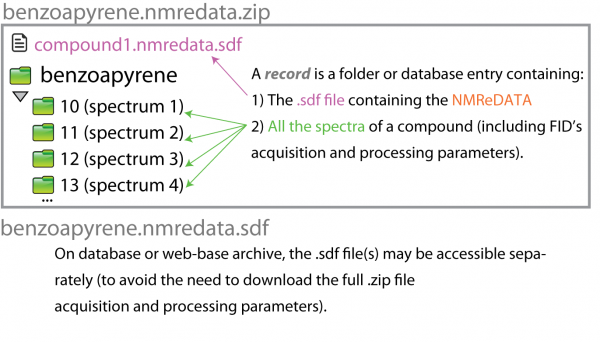
We call "NMR record", a folder (or .zip file including the folder) or a database record including:
1) All the NMR spectra (including FID, acquisition and processing parameters). The format of these data is as produced by the manufacturer of the instrument which acquired the data. That means that software generating the data either has these crude data available or it will ask the user to point to the crude data in order to include them in the NMR record.
2) The SDF file(s) including the NMReDATA (.nmredata.sdf file)
A more detailed picture (see File:Test-2.pdf) was presented in the poster presented in July at the Euromar 2017. Note: The NMREDATA tag "SIGNALS" was renamed "ASSIGNMENT" in Version 0.98.
NMR records will be requested by Magnetic Resonance in Chemistry from 2018 on. The editors of software (ADC/Labs, Bruker, cheminfo, Mestrelab) are able to grenerate MNR records.
Records will be either analysed on web pages, or downloaded, and the nmredata.sdf file opened by the software which will access automatically to the associated spectra.
The full description can be found in the NMReDATA tag format page.
An example (probably obsolete) of .nmredata.sdf files with the spectra can be found here
A better source is the GitHub example page of the GitHub page of the Initiative.
Mixtures of compounds
When more than one compound is present in a sample, multiple .sdf are produced and called compound1.nmredata.sdf, compound2.nmredata.sdf, etc.
Current version of the format of NMReDATA
The format can be found here : NMReDATA tag format
Various examples of NMReDATA files can be found in the github repository.
Database
The NMReDATA Inititiative define the format and rules for hosting and provide data, but hosting data is not part of our mission.
Discussion on about database providers
Versions
Changes to Versions 1.1
- Addition of backslash at the end of the line of NMReDATA tags.
Changes to Versions 2.0
- Possibility to include Jcamp spectra on top of original data from the instrument.
- Possibility to add a 3D structure (on top of the "flat" 2D structure).
- Possibility to add author and institution information (to facilitate integration of these metadata in repositories).
For details, see the specification.
Changes to Versions 2.1 (future version)
Future version of the format! Suggestions are welcome!
- Possibility to include J-coupling assignment data (coupling network and J-graph data).
Discussion and complete list on future versions of the NMReDATA format
Jmol integration in this wiki
NMReDATA is being integrated with Jmol/JspecView
Publications
- NMReDATA, a standard to report the NMR assignment and parameters of organic compounds. M. Pupier et al. (2018) Magnetic Resonance in Chemistry 56:703-715. doi: 10.1002/mrc.4737 PMID: 29656574
- NMReDATA: Tools and applications. S. Kuhn et al. (2021) Magnetic Resonance in Chemistry 59:792-803. doi: 10.1002/mrc.5146 PMID: 33729627