Main Page
Link to the MRC article on the NMReDATA format.
Direct link to page describing the format of the NMREDATA tags.
List of compatible software
Tentative instruction for journal submission of NMReDATA.
Discussion on future versions of the NMReDATA format
Contents
Introduction
The NMReDATA working group decided to include data extracted from NMR spectra of small molecules in SDF files using SD tags.
An important task of the group is to define the format of the content of the "<NMREDATA_...>" tags. More details here!.
The version 1.0 will be decided in September at the "Round table" of the Smash 2017 conference at Baveno, Italy.
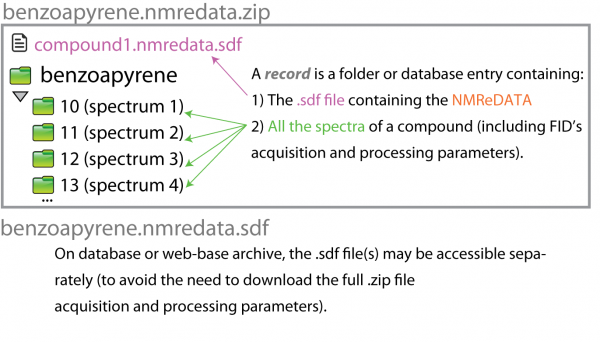
The SDF file alone (that is without the spectra) cannot be used to verify that the assignment corresponds to the spectra. It is therefore important to always have the spectra with the SDF file! We call "NMR Record" the combination of the spectra and the SDF file.
NMR records
We call "NMR record", a folder (or .zip file including the folder) or a database record including:
1) All the NMR spectra (including FID, acquisition and processing parameters). The format of these data is as produced by the manufacturer of the instrument which acquired the data. That means that software generating the data either has these crude data available or it will ask the user to point to the crude data in order to include them in the NMR record.
2) The SDF file including the NMReDATA (.nmredata.sdf file)
A more detailed pictorial representation of NMR record and example of SDF file presented in the poster presented in July at the Euromar 2017. Note: The NMREDATA tag "SIGNALS" was renamed "ASSIGNMENT" in Version 0.98.
NMR records will be requested by Magnetic Resonance in Chemistry from 2018 on. The editors of software (ADC/Labs, Bruker, cheminfo, Mestrelab) will be ready by the end of 2017 to produce MNR records for submission to MRC.
Records will be either analysed on web pages, or downloaded, and the nmredata.sdf file opened by the software which will access automatically to the associated spectra.
The full description can be found in the NMReDATA tag format page.
An example of .nmredata.sdf files with the spectra can be found here
Current version of the format of NMReDATA
The format can be found here : NMReDATA tag format
A small set of simple examples of .nmredata.sdf files can be found here (Version 0.95). (Note: the field called "NMREDATA_SIGNALS" was renamed "NMREDATA_ASSIGNMENT" in V 0.98). It includes .nmredata.sdf files for ethanol with diverse formats (explicit or implicit hydrogen atoms, etc.)
Versions
Changes to Versions 1.1
Addition of backslash at the end of the line in text of the NMReDATA tags. (This is to avoid the problem that CDK librairies ignore the NewLine character (ASCII 10). Since we need a line separator, we use backslash + NewLine in NMReDATA tags).
Comparison of assigned data
This is a working proposition
A tag comparing the data in the current file with those from other files (with the same labels for the assignement) > <NMREDATA_COMPARISON>
Externaldata1=... ref to an external .sdf file from the current or external record. Externaldata2=... H1 1.54 1.50 1.66 (the first value is the reference value in NMREDATA_ASSIGNMENT tag of the current file the second from Externaldata1, etc.) H2 1.54 1.50 1.66 Chi2 0.3 0.5 (these could be the chi squared of the reference with the external data )
Coupling could also be compared...
> <NMREDATA_COMPARISON>
Externaldata1=... ref to an external .sdf file from the current or external record. Externaldata2=... J(H1,H2) 1.54 1.50 1.66 (the first value is the reference value in NMREDATA_ASSIGNMENT tag of the current file the second from Externaldata1, etc.) J(H1,H3) 1.54 1.50 1.66 Chi2 0.3 0.5 (these could be the chi squared of the reference with the external data)
Other comparison could be made, for example from signal intensities in HSQC spectra
> <NMREDATA_COMPARISON>
Externaldata1=... ref to an external .sdf file from the current or external record. Externaldata2=... I(NMREDATA_13C_1J_1H,HC1,H1) 122 154 143 (the first value is the reference value in NMREDATA_ASSIGNMENT tag of the current file the second from Externaldata1, etc.) I(NMREDATA_13C_1J_1H,HC2,H2) 132 151 163 Chi2 5.3 3.5 (these could be the chi squared)
Certification
When the assignment is made using a computer-assisted manner, the software may want to add a certification of the validity of the data. This should be (up to the manufacturers) to somehow encode it to make it impossible to forge the certification (using hashtag, etc. ?) Certificates TAGS could be listed at the end of the .sdf file. They can originate from the CASE software or from the database hosting the data and spectra, from the journal (to say data were peer-reviewed). They can be cumulated. If the text of the .sdf file needs to be hashed for certification, the list of TAG used for hashing could be listed.
To be refined by the specialists!
> <NMREDATA_CERTIFICATION> Software=CMC_assist Author=Bruker Confidence_level=4.6 Confidence_level_certificate=”ADFS678AG67DFG6SD5F7GS5DFGSD8F5GSD7FG7” Unique_solution=YES Unique_solution_certificate=”ADFS678AG67DFG6SD5F7GS5DFGSD8F5GSD7FG7” etc.
This is only a very vague example. The uniqueness of the structure proposed may be understood in the sense of J.-M. Nuzillard’s Logic for Structure Determination (LSD) tool. Software producers can include here the specifications of their product. Multiple certification can be listed one after the other. The “Software=...” assignment separates them all in the same <CERTIFICATION> tag.
Role(s) and scope of the “assignment records”
The NMR record can be generated from experimental data (this is how the format was designed), but data may also originate from simulations, predictions, etc.
Tools to compare, evaluate, validate, and check consistency of “assignment records” will certainly be developed.
Assignment records can be generated by commercial software, but also by diverse tools analysing NMR data, homemade processing tools, simulation software, etc. This is why it is important to have a format of data including a maximum of options to be as flexible as possible, even if not all possible uses are clearly defined and used immediately. Ideally, the .sdf files should be converted into other file formats or spectral description without loss.
Multiple records
We should see as an advantage if databases include multiple "assignment records" associated to the same molecule or the same set of NMR spectra. Some could be old, originating from incomplete literature data. Others could include errors because they originate from bulk data processed automatically. But finally, a computer could verify or create a robustly validated record combining all the other data. Aggregated record could be generated by NMR software/database scoring available data for consistency, calculated chemical shifts and spectral simulations. They could refine chemical shifts and couplings, etc.
SDF files generated from experimental data
When the NMR data originate from experimental spectra, they may be quite crude (simple automated integration, peak-picking). At the other extreme, the data may follow complex automated or careful and manual expert analysis. The NMReDATA must have the flexibility to code diverse quality of data: They may be partial, incomplete, contain inconsistencies, impossible features, etc.
- only 1D 1H NMR data (with or without integration, coupling, etc.).
- only 1D 13C data (just from a simple peak peaking)
- only 1D data but for multiple isotopes (from NMRshiftDB ?)
- full analysis based on computer-assisted software (such as ACDLabs Structure Elucidator, Bruker CMC-se or Mestrelab Mnova) or web platform (such as cheminfo.org).
- 1D and 2D data processed automatically with ambiguities on the signal assignment and partial (for example not all signals are assigned) and/or ambiguous (due to lack of resolution, or other problems)
- The file may not contain the actual assignment, only the structure and the list of chemical shift (the assignment could be added by NMR tools).
- The data may come from a scientific report i.e. the text providing the description of the spectra.
Scripts could be written to convert such a "pure text" description into .sdf file and include the .mol file.
For assignment work made with only "paper and pencil", a tool allowing to draw a molecule, enter lists of signal names and 2D correlations could be easily made. We could consider to accept .pdf or pictures of the spectra when the original files do not exist anymore.
SDF files generated from calculated data
This is a working proposition to include JCAMP spectra in NMR Records / Discussion using Slack
The NMR data may originate from DFT calculations or any other type of predictor of chemical shifts, and/or coupling. In such a case, a general tag is added to provide information about the software. For example:
> <NMREDATA_ORIGIN> Source=Calculation method=DFT Geometry=method/basis set Shielding=method_basis set Coupling=method_basis set Software=... Version=...
References to addition files located in a folder called "DFT_GIAO_calculations" could be added. They could include: -the equation used to convert sheelding in chemical shifts (for 1H, 13C)
csH=((Sxx+Syy+Szz)/3-60)*0.98+5" csC=((Sxx+Syy+Szz)/3-130)*1.1+3.1" JHH=((???)/3-130)*1.1+3.1"
-the list of conformers calculated, their energies ?, the Boltzman ratio ?
-all 3D structures of the conformations in a conformations.sdf file (not the main NMReDATA .sdf file). The main .sdf file can contain the 3D structure of the lowest energy conformation and the flat structure only.
-all the output files of the shielding (and coupling) calculations ? Also the goemetry optimization ?
Examples (tentative) of .nmredata.sdf files originating from gaussian can be found here: androsten data
SDF files generated from literature data
This is a working proposition under review
When the NMR data originate from publications, a reference to the published paper/book/thesis is given in the NMREDATA_LITERATURE tag.
> <NMREDATA_LITERATURE> Source=Journal DOI=DOI_HERE (if Reference field is DOI specify it here) CompoundNumber=label used in the reference to designate the compound (typically a number in boldface)
> <NMREDATA_LITERATURE> Source=Book ISBN=ISBN_HERE (if Reference field is DOI specify it here) CompoundNumber=label used in the reference to designate the compound (typically a number in boldface)
> <NMREDATA_LITERATURE> Source=Thesis Thesis=HTML link here (if available if not "LastName, Firstname(s), institution providing the degree, city, country, year of publication. CompoundNumber=label used in the reference to designate the compound (typically a number in boldface)
SDF files generated after revision of existing SDF files
Assignment records may be generated after revision from experimental, literature, prediction data, etc. Ideally, the original .sdf files should also be generated to facilitate comparison or exist somewhere and be referred to. In both cases reference should be given.
> <NMREDATA_UPDATE> Source=Record Record_number=ref_to_the_original_record (multiple reference is allowed for aggregation of records – separated by “,”). Date =date.... standard format for date Correction="fixed assignments of C(13) and C(15)"
This is also to be refined according to future developments.
Concerning symmetry
Magnetic non-equivalence
For symmetrical molecules a difficulty may arise to code coupling and 2D correlations.
Reminder: Couplings are not directly associated to atoms, but to labels (in the NMREDATA_ASSIGNMENT tag). Labels are associated to one or more atoms (in case of symmetry/fast rotation, etc.).
Example of difficulty/solution concerning scalar coupling: For the 1H spectrum of 1, 2 dichlorobenzene, we have two multiplets in the 1D 1H spectrum (two different protons in an AA’XX’ system) so if the SDF file includes two labels (one for A and one for X, each pointing to two atoms), in principle one can only give one coupling: the JA,X (no JA,A or JA,X'). But if one desires to specify all the couplings, give two different "labels" to A and A' (each pointing to only one atom), so that different coupling can be given for JA,X, JA',X,JA,A', JX,X'. This may be desired so that the 1D spectrum can be simulated with the correct non-equivalence effect.
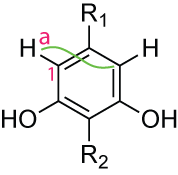
HMBC correlations in symmetrical molecules
Consider the following molecules:
A 3JC,H HMBC correlation will be visible between the proton a and C(1) that seems to be the directly-bound carbon. Because the carbons 1J and 3J bond, relative to a proton are symmetrical. A software may see the correlation as 1J, but, it should be able to analyse the NMREDATA_ASSIGNMENT tag and see that a and C(1) are pointing to two atoms, and that the correlation may correspond to any combination of the four possible pairs. Two pairs will seem as the actual 3J and two as the 1J.
Why not add more data in NMReDATA tags?
We consider that our task is to focus on NMR data. But SDF files could (and probably should!) also include other experimental data such as:
1) The origin of the molecule. This may include the extraction method and the plant it originates from, in phytochemistry, or the reaction producing it.
2) MS data
3) other spectral data
In principle authors can add any tag provided they have tools to do it and requests from the Journals... such data could have the following form...
The software producing SDF files including NMReDATA, should read SDF files and write SDF files only adding (or modifying/reviewing) the NMReDATA data. Any other SDF tags should be passed from the file which is read to the file which is generated.
THIS SITE HAS BEEN VANDALIZED. SINCE THEN, AS A PRECAUTION, I BLOCKED CONFIRMED USERS TO MODIFIY THE CONTENT OF THIS SITE AND ADD NEW PAGES.
PLEASE LET ME KNOW BY E-MAIL IF/WHEN YOU WANT TO MAKE CHANGES TO THIS WIKI SO THAT I ALLOW (TEMPORARILY) MODIFICATIONS AGAIN.